Google-owned DeepMind AI reveals 3D structures of 'entire protein universe'
What's the story
It took Ferdinand Perutz and John Kendrew years of work to finally unlock the structure of globular proteins. Over six decades later, AlphaFold, the revolutionary AI network of Google's London-based AI company DeepMind, has predicted structures for nearly all cataloged proteins known to mankind in just about 12 months. Do you know how many that is? Over 200 million proteins.
Context
Why does this story matter?
The subject of artificial intelligence can still easily cause a heated discussion. Both proponents and opponents have strong arguments to support their cases. However, we can all agree that artificial intelligence can certainly accelerate scientific discoveries. DeepMind's AlphaFold is a prime example of that. Its benefits outweigh the risks by a huge margin, and the new achievement is a testament to that.
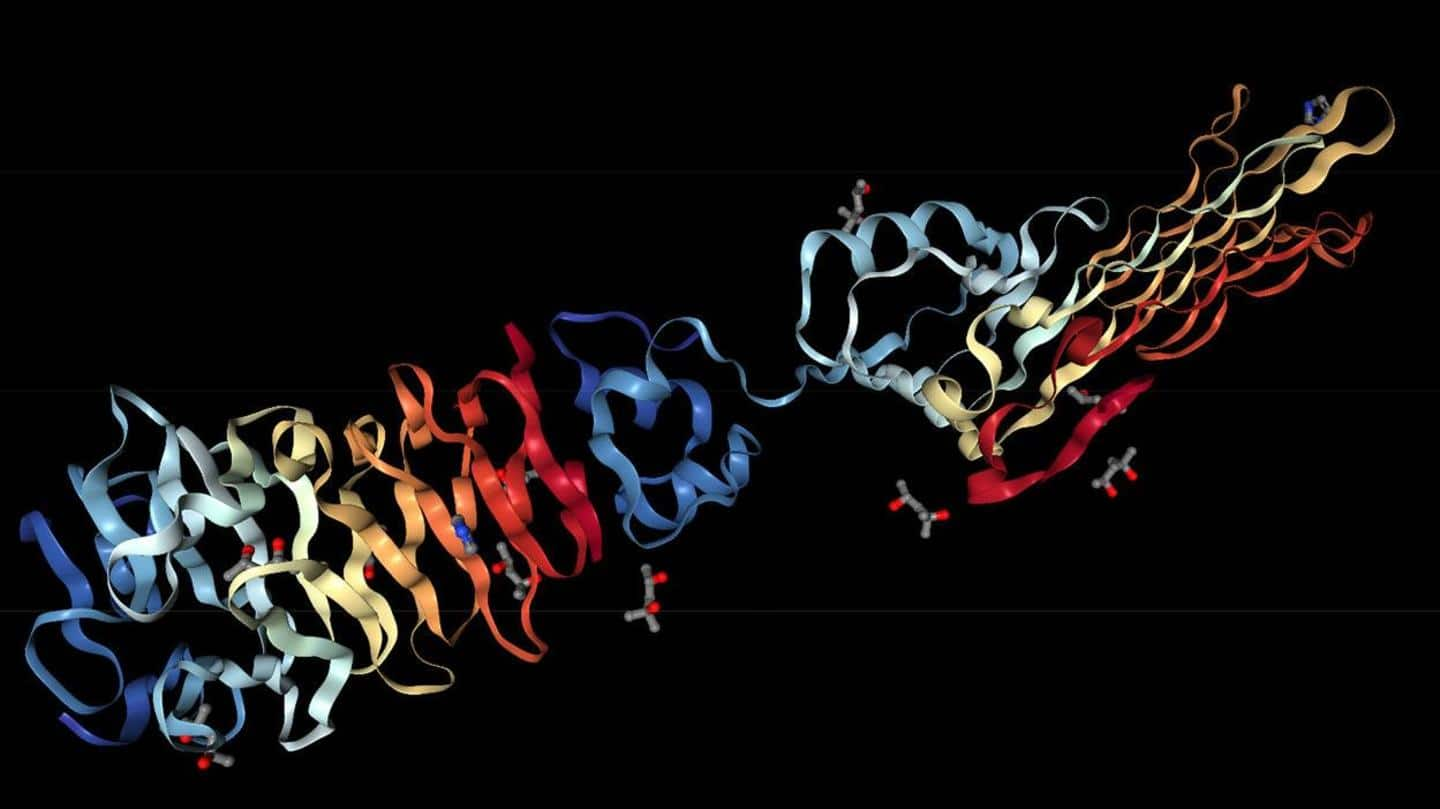
3D protein
AlphaFold can predict 3D structures from chemical composition
AlphaFold uses a technique called 'deep learning' to determine the 3D structure of proteins. It can predict the structure of a protein from its chemical composition. When it was launched a year ago, it had a database of 350,000 structures, which is now at a mind-boggling 200+ million. The database is set up in partnership with European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI).
Accuracy
How accurate are AlphaFold's predictions?
Traditionally, scientists have used methods such as X-ray crystallography and cryo-electron microscopy, which are both time-consuming and expensive to understand protein structures. AlphaFold made it easier for the scientific community by predicting highly accurate structures with much fewer hassles. According to EMBL-EBI, around 35% of predictions are deemed highly accurate, while another 45% are considered good enough to rely on for many applications.
Impact
The AI network's code and data are free to use
Since its introduction, AlphaFold has proved to be a major blessing for scientists. DeepMind's CEO Demis Hassabis described it as a "gift" to humanity as it is free to use for any purpose and has an irrevocable open source license. According to Hassabis, over 500,000 scientists have used the database to view over two million structures. It has helped scientists make new discoveries.
Information
AlphaFold's database has 23 terabytes of content
For its predictions, AlphaFold used another database called UniProt. Now, every protein sequence on UniProt will have a corresponding structure. AlphaFold's database is now 23 terabytes big and all 200+ million structures can be downloaded via Google Cloud Public Datasets.
Complication
Understanding the structure of proteins is important for developing medicines
Understanding the structure of proteins is important for developing medicines and in other areas of biochemistry. It is the interplay between molecules and proteins that gives the desired result. However, this process is complicated due to the structural complexity of proteins. Although they are amino acid strings, they can fold, bend, and even get tangled. In fact, a protein's folds determine how it functions.
Importance
AlphaFold can solve 'protein folding' issues faced by scientists
Scientists spend countless hours understanding the structure of proteins so that their folds don't block molecules from attaching. In the case of drugs, any mistake can lead to adverse reaction. AlphaFold is seen as a solution to this 'protein folding problem.' Its huge database can help scientists understand protein structure without spending a lot of time in labs and in an economical way.
Examples
Several scientific experiments and articles have used AlphaFold's structures
Since its introduction, AlphaFold has driven several scientific endeavors. University of Yale scientists used the database to help in their goal to create a highly effective Malaria vaccine in April. Similarly, a University of California experiment to understand the coronavirus and prepare for other pandemics received a much-needed push with AlphaFold's predictions. There have also been thousands of scientific articles that used AlphaFold's structures.